Doping editor
1. Overview
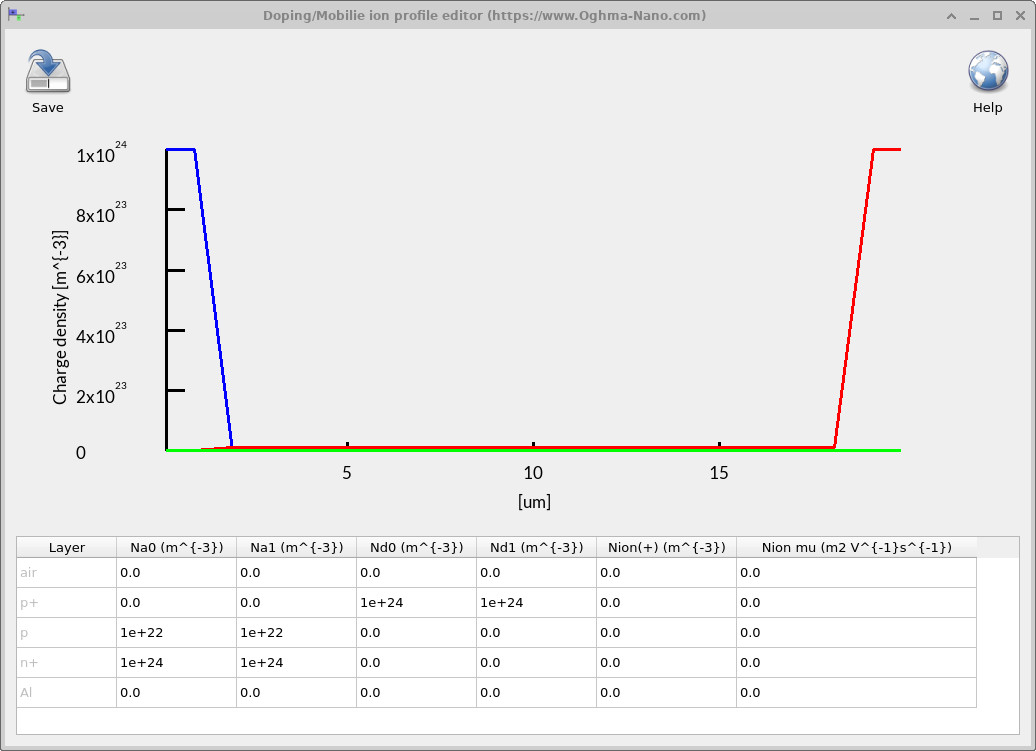
Use the Doping editor to set fixed ionized dopants (acceptors/donors) in each layer and, if relevant, add mobile ions for perovskites. Open it from the Electrical ribbon by clicking the Doping/Ions icon. The plot shows the resulting profiles across the device; the table beneath is where you edit numeric values.
2. Parameters
The table exposes the following fields (units in SI):
- Layer — layer name (read-only for display/context).
- Na0, Na1 (m-3) — ionized acceptor density at the start and end of the layer. Setting different values produces a linear grade through the layer thickness.
- Nd0, Nd1 (m-3) — ionized donor density at the start and end of the layer. Again, different values create a linear grade.
- Nion(+) (m-3) — density of mobile positive ions (for perovskites and similar systems). This value is uniform per layer (no grading).
- Nion μ (m2 V-1 s-1) — mobility of the mobile ions in that layer.
The graph updates to reflect the specified profiles. Acceptors/donors can be linearly graded within each layer using the 0/1 start–end pairs; mobile ions are not graded and are treated as a constant per layer with the specified mobility.
3. How they are applied
Fixed dopants contribute to the space charge in Poisson’s equation and set built-in fields, depletion/accumulation at interfaces, and the equilibrium carrier landscape. Linear grading within a layer creates internal electric fields that influence drift–diffusion transport and hence JV characteristics.
When mobile ions are present, their density and mobility determine how charge redistributes under bias and illumination. This affects transients (e.g., slow hysteresis and relaxation) and steady-state operating points in perovskite devices. If mobile ions are not relevant for your stack, leave Nion(+) at zero.